A well-planned experiment, with appropriate controls, treatments, and conditions, is often the first step toward obtaining improved results. To learn more about planning your western blot experiments, check out our Western Blotting Experimental Guidelines.
Select a troubleshooting topic of interest:
| Problem | Possible Causes | Recommendation |
|---|---|---|
|
Low or No Signal |
Reusing Pre-diluted Antibodies |
Reusing diluted antibody is not recommended because the antibody is less stable after dilution and older dilution buffer is prone to microbial or fungal contamination. We recommend always using freshly diluted antibody for optimal results. |
|
Low Protein Expression in Tissue or Cell Line |
We recommend using expression profiling tools such as BioGPS or The Human Protein Atlas as well as scientific literature to check whether or not your cells or animal tissues are expected to sufficiently express the target protein of interest. We always recommend including a known positive control to confirm experimental results. A list of recommended controls for many of our antibodies can be found on our Control Treatments by Target table. A protein load of at least 20-30 ug per lane is recommended for whole cell extracts and for detection of total/unmodified targets in whole tissue extracts. However, it is often necessary to increase the total protein load to at least 100 ug per lane for detection of modified targets (e.g., phosphorylated and cleaved) in whole tissue extracts. A higher protein load for whole tissue extracts may be necessary when only a small portion of the cells in the tissue includes the post-translationally modified target. Inclusion of protease and phosphatase inhibitors in the cell extract is essential to avoid protein degradation and maintain protein yield. We recommend that the lysis buffer include leupeptin (1.0 ug/ml final concentration) and PMSF (#8553) as protease inhibitors. Protease Inhibitor Cocktail (100X) (#5871) or Protease/Phosphatase Inhibitor Cocktail (100X) (#5872) may also be used. |
|
|
Low Level of Phosphorylated or Modified Protein |
Many post-translationally modified proteins are expressed at low basal levels in cell lines or tissues without treatment. We recommend using PhosphoSitePlus to look up low-throughput papers referencing your particular modification site or use our Control Treatments by Target table to find an example of a treatment and cell line or tissue that works well as a positive control. Inclusion of protease and phosphatase inhibitors in the cell extract is essential to avoid protein degradation and maintain protein yield. Sodium pyrophosphate (2.5mM final concentration) and beta-glycerophosphate (1.0mM final concentration) should be included as serine/threonine phosphatase inhibitors in the lysis buffer. Sodium orthovanadate (2.5 mM final concentration) should be included to inhibit tyrosine phosphatases. Protease Inhibitor Cocktail (100X) (#5871) or Protease/Phosphatase Inhibitor Cocktail (100X) (#5872) may also be used. 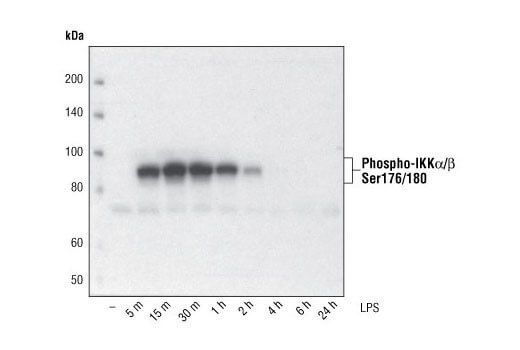
|
|
|
Secreted Proteins |
Some target proteins are secreted from the cell and cannot be reliably detected in whole cell extract. Cell media can be precipitated using acetone or concentrated to detect the secreted target. In some cases, chemical modulators such as Brefeldin A (#9972) can be used to inhibit the secretion of your protein of interest from the cell. 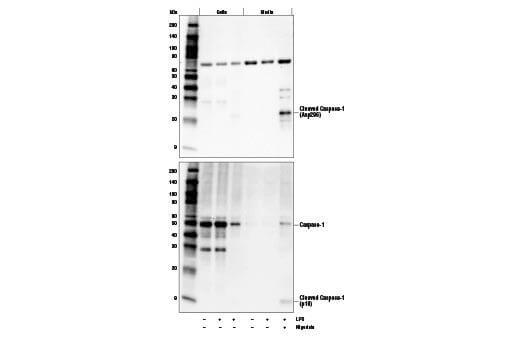
|
|
|
Antibody Sensitivity and Reactivity |
Be sure to check the Sensitivity/Specificity section of the antibody's webpage. A transfected- or recombinant-only antibody is not sensitive enough to detect its target at endogenous levels. Endogenous sensitivity antibodies will work with all sample types (endogenous, transfected, or recombinant protein). The amount of antibody used may also need to be increased or decreased to achieve optimal signal depending on the abundance of your target protein in your samples. We recommend using the dilution shown on the product webpage and datasheet as a starting point for optimization. Another factor to consider is the species reactivity of the antibody. Species listed in the Reactivity section on a product webpage have been validated in-house by CST. If your species of interest is not listed, please contact our Technical Support team and they will be happy to help determine the predicted reactivity of the antibody with the protein sequence of your model species. |
|
|
Incomplete Lysis |
CST recommends sonication to ensure complete lysis and maximal/consistent protein recovery regardless of the lysis buffer used. Sonication is essential to ensure efficient protein extraction of membrane-bound and organelle localized (e.g., nuclear and mitochondrial) targets and also serves to shear nuclear DNA that can interfere with the loading of SDS-PAGE gels. Probe sonicators are recommended for the best sample preparation. For 1 mL samples, 3 x 10 second bursts with a microtip probe sonicator at 15W (or 50% power) on ice works best. However, good alternatives are repeated passage through a fine gauge (e.g., 24 gauge) needle or vortexing with glass beads. After sonication, the sample can be centrifuged to pellet insoluble cellular debris and the supernatant used for western blotting. |
|
|
Sub-optimal Buffer Choice |
For optimal results with individual antibodies, please consult the western immunoblotting protocol on the product webpage for the recommended primary antibody dilution buffer (i.e., BSA or milk). Failure to incubate our antibodies in the recommended dilution buffers can severely compromise the sensitivity/specificity of the results. For example, nonfat dry milk is too stringent for primary antibody incubations with many of our antibodies. In general, CST recommends the blocking and secondary antibody incubations be performed using 1X TBS/0.1% Tween-20/5% nonfat dry milk in order to minimize non-specific background. The use of alternate blocking agents, such as gelatin, serum, protein-free blocking agents, casein, or mixed blocking agents may reduce target signal intensity. Many customers are concerned about the use of non-fat dry milk as a dilution buffer for the detection of phospho-proteins. Specifically, it is believed that the casein in milk leads to high background. At CST, we have not observed any problems when using non-fat dry milk for the detection of phospho-proteins. All CST antibodies are tested in both 5% non-fat dry milk and 5% BSA prior to release and the optimal dilution buffer is chosen for recommendation. The blocking, antibody incubation (primary and secondary), and washing buffers should all include 1X TBS/0.1% Tween-20. A higher or lower percentage of Tween-20 can compromise the sensitivity/specificity of the results. The use of PBS rather than TBS may weaken the signal intensity with some of our antibodies. 
|
|
|
Sub-optimal Transfer Conditions |
We typically recommend wet transfers at 4°C for 2 hours at 70V (200-250mA) in 25mM Tris, 192mM Glycine, and 20% methanol. However, for high molecular weight proteins, we recommend decreasing the methanol content of the transfer buffer to 5-10% and increasing the transfer time to 3-4 hours (200-250mA) at 70V. For semi-dry transfer, please refer to the system manufacturer's recommendations. For low molecular weight proteins, it is important to avoid over transfer or "blow through" with smaller targets. We recommend a shorter transfer time and using nitrocellulose transfer membrane with a pore size of 0.2 um to minimize the loss of low molecular weight proteins (i.e., less than 25-30 kDa). |
|
|
Multiple Bands or Non-specific Binding |
Isoform Reactivity |
Some cell line and tissue models can contain more than one protein isoform or splice variant, which can migrate at different molecular weights. Refer to the Specificity / Sensitivity section on the antibody's webpage to determine if it is predicted or confirmed to detect more than one isoform. You may also reference your protein of interest on UniProt to see if multiple isoforms sequences are listed. |
|
Post-translational Modifications |
Post-translational modifications (PTMs) may cause a subset of the target protein to run at a different rate than the unmodified protein. Therefore, treatment conditions that induce or inhibit certain post-translational modifications can lead to multiple bands being observed on a western blot. Glycosylation, SUMOylation, ubiquitylation, and phosphorylation are examples of modifications that can cause multiple bands to appear on a western blot, depending on the samples and treatments used. If you are looking for more information on PTMs associated with your target protein, please see PhosphoSitePlus. |
|
|
Tissue Samples |
Because tissue samples are non-homogenous (i.e., composed of different cell types), the chances of non-specific bands appearing with an antibody that may not be detected in other sample types such as cell lines is increased. In addition, some tissue models may contain more than one protein isoform or splice variant, which can migrate at different molecular weights. |
|
|
Lysate Degradation |
Proteases and phosphatases in your sample can quickly degrade your protein. Adding protease and phosphatase inhibitors to your lysate can help slow or prevent protein degradation. We recommend the lysis buffer include leupeptin (1.0 ug/ml final concentration) and PMSF (#8553) as protease inhibitors. Protease Inhibitor Cocktail (100X) (#5871) or Protease/Phosphatase Inhibitor Cocktail (100X) (#5872) may also be used. The age of a lysate can also lead to an increase in protein degradation products, which are then picked up by the antibody. Ensure you are using a fresh sample to minimize protein degradation. Some proteins are less stable than others in the same vial of sample, so although a sample may work for one protein target, a different protein may already be degrading in the same sample. 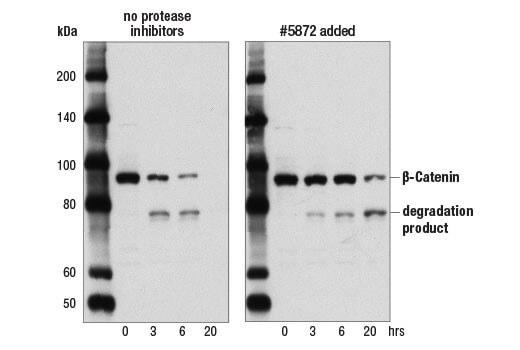
|
|
|
High Protein Concentration |
If working with a highly sensitive antibody, excess protein on the membrane can cause the appearance of multiple bands and high background as well as intense signal throughout the lane that may not otherwise be there with a smaller amount of protein. Try loading less protein to observe a cleaner signal. |
|
|
Long Exposure Time |
If the amount of protein loaded is too low, it will make it difficult to detect your protein of interest, especially if it is less abundant in your sample type. This in turn will lead to a need to increase the exposure time to detect the target protein on your blot. CST antibodies are formulated to produce signal within a two-minute exposure time with common ECL reagents. If no signal is seen within this time, a higher protein concentration may be needed. |
|
|
Sub-optimal Primary Antibody Dilution Buffer |
The buffer used for the dilution of the primary antibody can affect your membrane background. As an example, a buffer such as non-fat dry milk will do a better job at decreasing the appearance of non-specific bands and overall background on the blot, compared to BSA. However, milk may be too stringent for some antibodies and lead to reduced target signal. It is important to consult the recommended western immunoblotting protocol on the CST product webpage to choose the appropriate primary antibody dilution buffer (i.e., BSA or nonfat dry milk). In general, CST recommends the blocking and secondary antibody incubations be performed using 1X TBS/0.1% Tween-20/5% nonfat dry milk in order to minimize non-specific background. 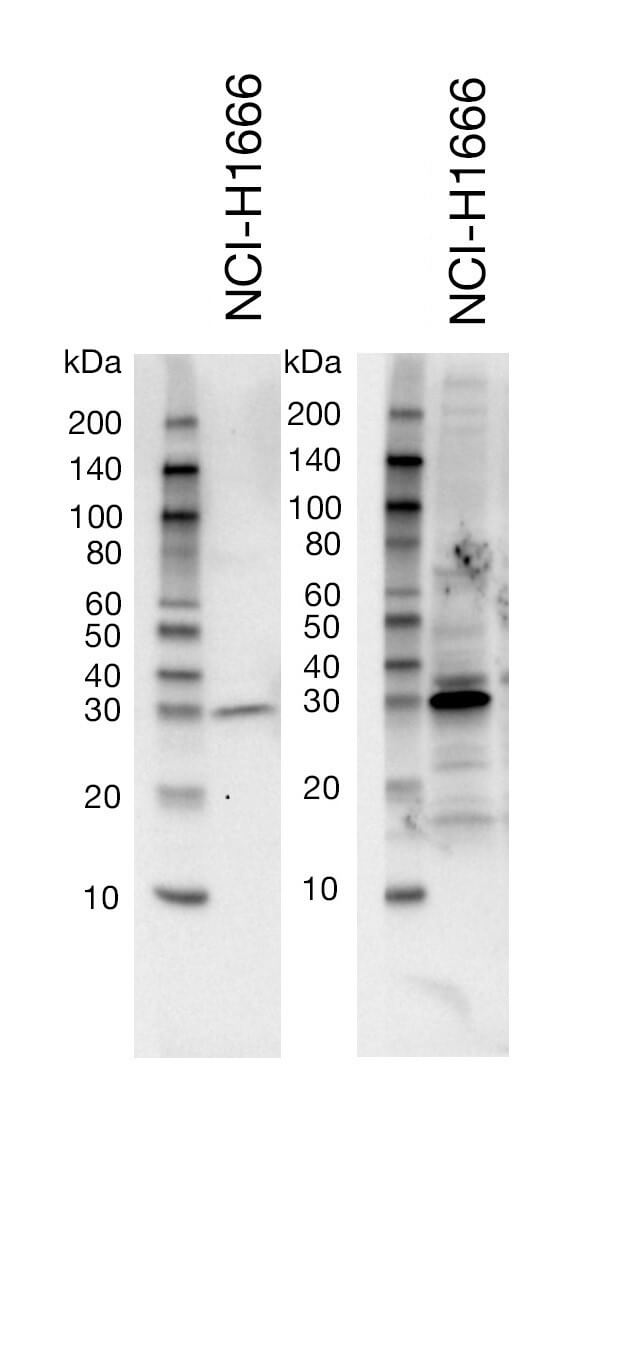
|
|
|
Smearing |
Glycosylated Proteins |
Differential glycosylation often results in a smear of bands at higher than predicted molecular weights. Potential sites for glycosylation in your target protein can be found at PhosphoSitePlus. Treatment of your samples with PNGase F, an enzyme that cleaves N-glycans from glycoproteins, can also be used to confirm a shift in molecular weight due to glycosylation. 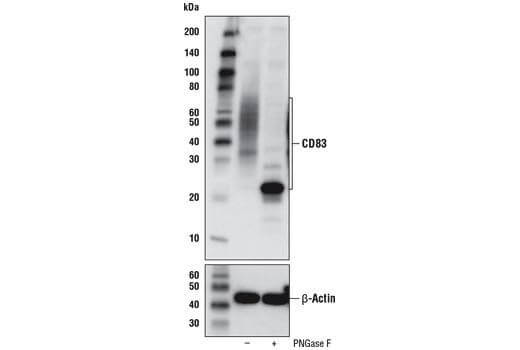
|
|
Lysate Degradation |
Proteases and phosphatases in your sample can quickly degrade your protein. Adding protease and phosphatase inhibitors to your lysate can help slow or prevent protein degradation. We recommend the lysis buffer include leupeptin (1.0 ug/ml final concentration) PMSF (#8553) as protease inhibitors. Protease Inhibitor Cocktail (100X) (#5871) or Protease/Phosphatase Inhibitor Cocktail (100X) (#5872) may also be used. The age of a lysate can also lead to an increase in protein degradation products, which are then picked up by the antibody and can appear as a smear below the expected target molecular weight. Ensure you are using a fresh sample to minimize protein degradation. If long-term storage is necessary, we recommend storing lysates at -80°C to reduce degradation. Some proteins are less stable than others in the same vial of sample, so although a sample may work for one protein target, a different protein may already be degrading in the same sample. |
|
|
Incomplete Lysis |
CST recommends sonication to ensure complete lysis and maximal/consistent protein recovery regardless of the lysis buffer used. Sonication is essential to ensure efficient protein extraction of membrane-bound and organelle localized (e.g., nuclear and mitochondrial) targets and also serves to shear nuclear DNA that can interfere with the loading of SDS-PAGE gels. Probe sonicators are recommended for the best sample preparation. For 1 mL samples, 3 x 10 second bursts with a microtip probe sonicator at 15W (or 50% power) on ice works best. However, good alternatives are repeated passage through a fine gauge (e.g., 24 gauge) needle or vortexing with glass beads. After sonication, the sample can be centrifuged to pellet insoluble cellular debris and the supernatant used for western blotting. |
|
|
Dark or Black Blot |
High Concentration of Secondary Antibody |
Black blots with or without white, “ghost” bands most often result when too much secondary antibody (i.e., horseradish peroxidase) is used with more sensitive detection reagents [e.g., our SignalFire™ Elite ECL Reagent (#12757)]. This is best addressed by increasing the dilution of the HRP-conjugated secondary antibody (e.g., from 1:2000 to 1:10,000). 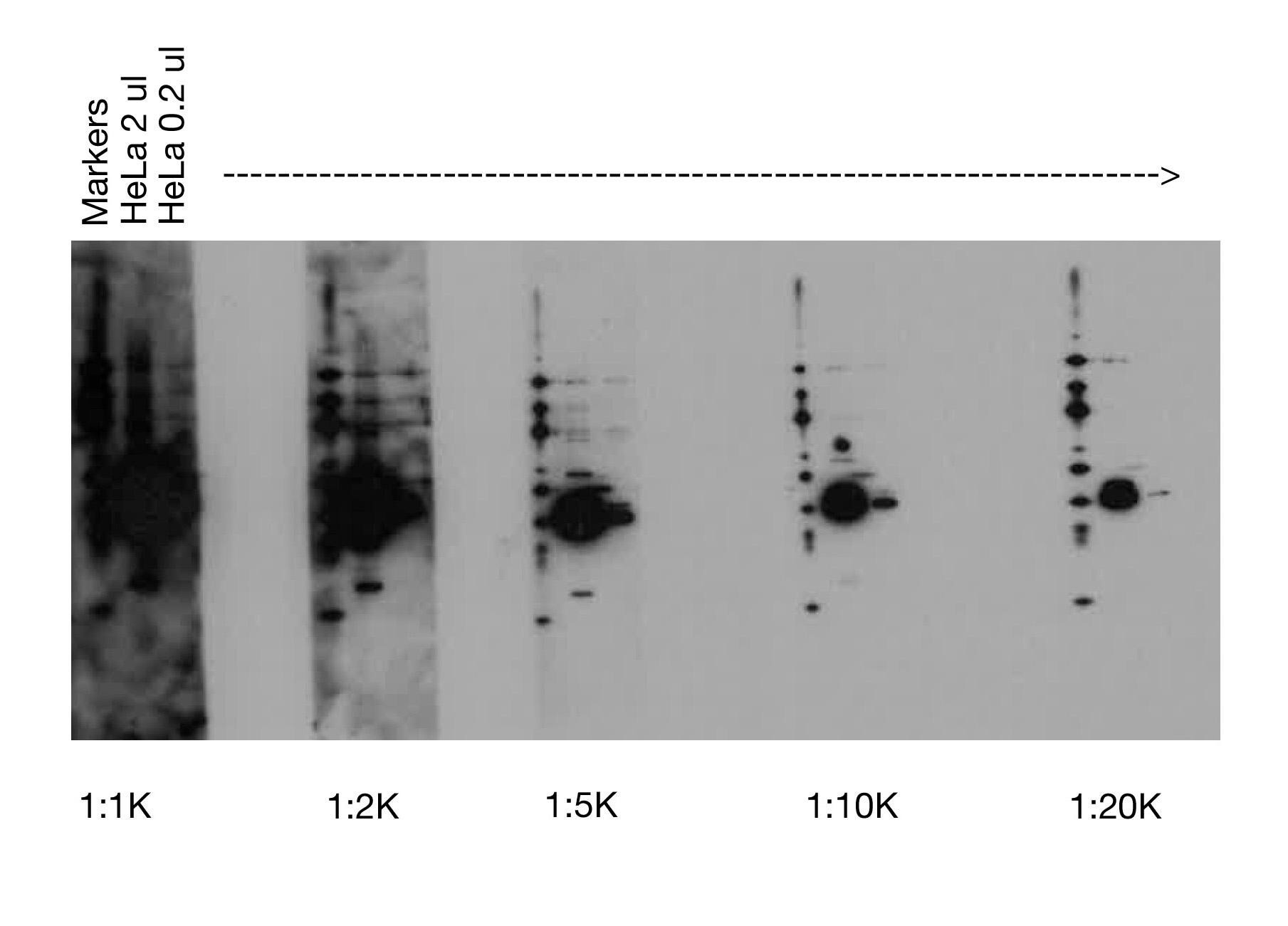
|
|
Insufficient Washing |
Reduced wash times may lead to excess antibody being left on the membrane, causing a dark blot after chemiluminescent exposure. CST recommends performing three, five-minute washes at room temperature in 1X TBS/0.1% Tween-20 after both the primary and secondary antibody incubations for optimal results. |
|
|
Sub-optimal Buffer Choice |
The blocking buffer used, and inclusion or absence of additional detergent can affect your membrane background. As an example, a buffer such as non-fat dry milk will do a better job at decreasing the appearance of non-specific bands and overall background on the blot, compared to BSA. However, milk may be too stringent for some antibodies and lead to reduced target signal. It is important to consult the recommended western immunoblotting protocol on the product webpage to choose the appropriate dilution buffer for the primary antibody incubation (i.e., BSA or nonfat dry milk). In general, CST recommends the blocking and secondary antibody incubations be performed using 1X TBS/0.1% Tween-20/5% nonfat dry milk to minimize non-specific background. |
|
|
Membrane Rehydration |
For optimal performance, we recommend rehydrating nitrocellulose membranes directly in the transfer buffer. However, when working with PVDF, the membrane should be briefly rehydrated in methanol before moving the membrane to the transfer buffer. |
|
|
Speckled or Splotchy Blot |
Contaminated Transfer Sponges |
Older transfer sponges that have not been properly sanitized may harbor bacterial contaminants. To rule out this factor, use new transfer sponges. |
|
Contaminated Buffers |
Buffers older than a few days will contain increased bacterial and fungal contaminants that can lead to speckling on the membrane. We recommend always using freshly made buffers for optimal results. |